***.cirpol
***.000
To combine the tilt images and to rematch all images, NIH Image and the Lazy stack macro are used. First, the variously focussed tilt images are combined into one for each tilt. Next, the circular polarization image (1), the rotation images (19) and the tilt images (2) are combined to a stack.
For better overall focus, the tilt images are combined from two to three images which are taken with varying focus levels. Using NIH Image, the two or three images of a given tilt are combined to a stack and the appropriate half or third of each slice is copied and pasted to the other slice(s) thus combining the focussed parts of the separate images into one. Key f and g of the Lazy stack macro can be used to select the appropriate regions of interest (above or to the right of the cursor position). In this process of combining variously focused tilt image into one, distortion is actually reduced.
The images (1 cirpol, 19 rot's, 2 tilt's) are opened and combined to a stack. It is useful to stick to a given file order (by given alphabetical names), as the stack management of NIH Image does not retain the original file names of the slices. Lazy stack macro provides a number of procedures to rematch the slices.
|
|
|
|
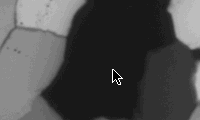
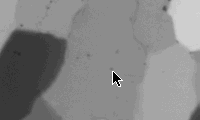
***.cirpol |
***.000 |
On the first slice (***.cirpol), find a black grain (=> c-axis normal to thin section), go to next slice (***.000): the black grain appears grey. Dust specks on this grain are suited as marker point as they will appear on all rotation and tilt images. Press ða (select all), and position the tip of the arrow next to the speck (see figure). Press ð. (period) to go to next slice: the marker point is probably NOT at the tip of the arrow. Press ðcðv (copy, paste), press ÆØ..etc. (arrow keys) to move the marker point to the tip of the arrow. If the marker point is more than 10 pixels away from the tip of the arrow, press 5, 6, 7, 8 to move ten pixels right, down, left and up respectively. Press ða (select all) BEFORE you move to next slice (or you loose the arrow). Save matched stack.
The images of the stack overlap only in the central part. Use s to select a 786·626 region of interest (ROI) near the center. By paging through the stack verify if the ROI is in the image area of all slices. If necessary move ROI accordingly. Use 1, and scale factor 1.00, to crop the stack. If necessary, this stack can be rematched again. Use s again to select a 780·620 ROI in the center of this stack and using 1, and scale factor 1.00, to crop. Save stack.
Use NIH Image menu Stacks to convert the 780·620 stack to windows. Note that these windows do not have names; they are numbered and appear cascaded from 001 to 024 (=top image).
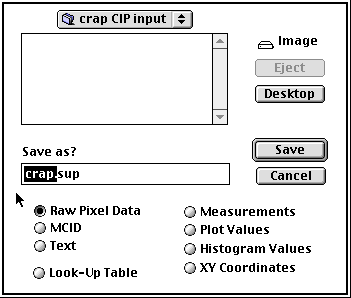
Use $ to export top image - as Raw Pixel Data - and save it in the appropriate CIP input folder. Type a file name and the extension ".sup" into the filename selection box, and copy the filename. Press return, the image is saved and closed. Use $ again, press ðv to copy the filename into the filename selection box, and type file name extension "eup" by hand. Continue to save slices backwards; the last slice, image no. 001, should be "cirpol".